nmat1390[1] Quantum dot bioconjugates for imaging, labelling and sensing.pdf
- 文件大小: 1.63MB
- 文件类型: pdf
- 上传日期: 2025-08-18
- 下载次数: 0
概要信息:
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 435
Quantum dot bioconjugates for imaging,
labelling and sensing
One of the fastest moving and most exciting interfaces of nanotechnology is the use of quantum
dots (QDs) in biology. The unique optical properties of QDs make them appealing as in vivo and
in vitro fl uorophores in a variety of biological investigations, in which traditional fl uorescent labels
based on organic molecules fall short of providing long-term stability and simultaneous detection of
multiple signals. The ability to make QDs water soluble and target them to specifi c biomolecules has
led to promising applications in cellular labelling, deep-tissue imaging, assay labelling and as effi cient
fl uorescence resonance energy transfer donors. Despite recent progress, much work still needs to be
done to achieve reproducible and robust surface functionalization and develop fl exible bioconjugation
techniques. In this review, we look at current methods for preparing QD bioconjugates as well as
presenting an overview of applications. The potential of QDs in biology has just begun to be realized
and new avenues will arise as our ability to manipulate these materials improves.
IGOR L. MEDINTZ1*, H. TETSUO
UYEDA2, ELLEN R. GOLDMAN1 AND
HEDI MATTOUSSI2*
1Center for Bio/Molecular Science and Engineering, Code 6900,
US Naval Research Laboratory, Washington, DC 20375, USA
2Division of Optical Sciences, Code 5611, US Naval Research
Laboratory, Washington, DC 20375, USA
*e-mail: Imedintz@cbmse.nrl.navy.mil;
Hedimat@ccs.nrl.navy.mil
One of the fundamental goals in biology is
to understand the complex spatio–temporal
interplay of biomolecules from the cellular to
the integrative level. To study these interactions,
researchers commonly use fluorescent labelling
for both in vivo cellular imaging and in vitro
assay detection1. However, the intrinsic photo-
physical properties of organic and genetically
encoded fluorophores, which generally have
broad absorption/emission profiles1 (Fig. 1a)
and low photobleaching thresholds, have limited
their effectiveness in long-term imaging and
‘multiplexing’ (simultaneous detection of multiple
signals) without complex instrumentation and
processing2. The seminal publications describing
the first biological uses of QD-fluorophores
foresaw that their unique properties could
overcome these issues3,4. QD properties of interest
to biologists include high quantum yield, high
molar extinction coefficients (~10–100× that of
organic dyes)5,6, broad absorption with narrow,
symmetric photoluminescence (PL) spectra
(full-width at half-maximum ~25–40 nm)
spanning the UV to near-infrared (Fig. 2), large
effective Stokes shifts (Fig. 1b,c), high resistance
to photobleaching and exceptional resistance to
photo- and chemical degradation (Fig. 3a)7–11.
Compared with molecular dyes, two properties
in particular stand out: the unparalleled ability
to size-tune fluorescent emission as a function of
core size (for binary semiconductor materials),
and the broad excitation spectra, which allow
excitation of mixed QD populations at a single
wavelength far removed (>100 nm) from
their respective emissions (Fig. 1b,c). QDs
also display intermittency (blinking) under
continuous excitation, a property only partially
understood, and which has been attributed to
Auger ionization12,13.
SYNTHESIS AND CAPPING STRATEGIES
QDs used in bio-applications are exclusively
colloidal nanocrystals. The best available QD
fluorophores for biological applications are made
of CdSe cores overcoated with a layer of ZnS
because this chemistry is the most refined. The
ZnS layer passivates the core surface, protects it
from oxidation, prevents leeching of the Cd/Se
into the surrounding solution and also produces a
substantial improvement in the PL yield5,14. Even
though thin ZnS (1–2 monolayers) shells often
nmat1390-print.indd 435 11/5/05 10:18:11 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
436 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
produce the highest PL yields, thicker ZnS shells
(4–6 monolayers) provide more core protection
against oxidation and the harsher conditions
presented by biological media (for example, acidic
buffers and cellular organelles). Colloidal QDs
made of ZnS, CdS, ZnSe, CdTe and PbSe, emitting
from the UV to the infrared have been prepared14–
29 (Fig. 2); however, these may need refinement
for bio-applications as issues of reproducible
synthesis and inorganic passivation remain.
The real breakthrough in synthesizing high-
quality colloidal QDs came when the Bawendi
group reported24 that use of high-temperature
growth solvents/ligands (mixture of trioctyl
phosphine/trioctyl phosphine oxide, TOP/TOPO),
combined with pyrolysis of organometallic
precursors, yielded CdSe QDs with highly
crystalline cores and size distributions of 8–11%.
The same reaction combined with appropriate
organometallic precursors was further used to
1.0
0.5
0.0
1.0
0.5
0.0
1.0
0.5
0.0
1.0
0.5
0.0
Ab
so
rb
an
ce
(n
or
m
al
iz
ed
)
Ab
so
rb
an
ce
(n
or
m
al
iz
ed
)
Photolum
inescence (norm
alized)
Photolum
inescence (norm
alized)
Absorption rhodamine red
Absorption DsRed2
Emission rhodamine red
Emission DsRed2
510 nm ABS
510 nm EM
530 nm EM
555 nm EM
570 nm EM
590 nm EM
610 nm EM
400 450 500 550 600 650 700
Wavelength (nm)
rCdSe core (Å)
λmax em. (nm)
13.5
510
14.5
530
17.5
555
19.0
570
21.5
590
24.0
610
b
a
c
d
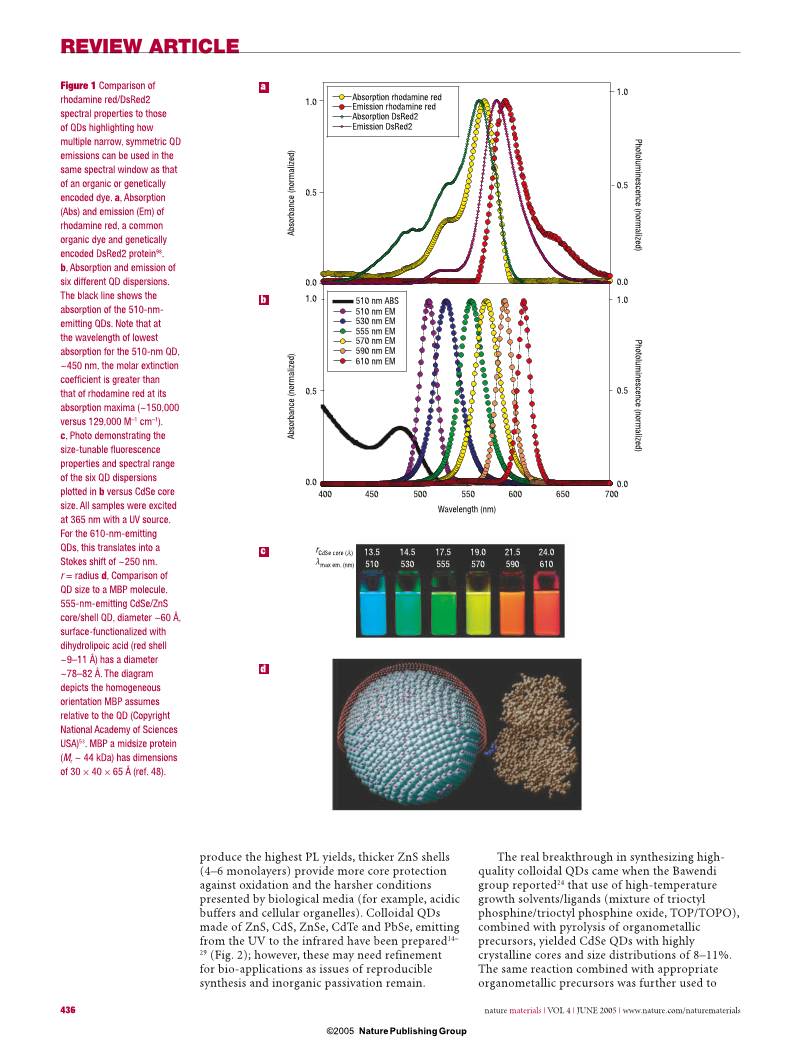
Figure 1 Comparison of
rhodamine red/DsRed2
spectral properties to those
of QDs highlighting how
multiple narrow, symmetric QD
emissions can be used in the
same spectral window as that
of an organic or genetically
encoded dye. a, Absorption
(Abs) and emission (Em) of
rhodamine red, a common
organic dye and genetically
encoded DsRed2 protein98.
b, Absorption and emission of
six different QD dispersions.
The black line shows the
absorption of the 510-nm-
emitting QDs. Note that at
the wavelength of lowest
absorption for the 510-nm QD,
~450 nm, the molar extinction
coeffi cient is greater than
that of rhodamine red at its
absorption maxima (~150,000
versus 129,000 M–1 cm–1).
c, Photo demonstrating the
size-tunable fl uorescence
properties and spectral range
of the six QD dispersions
plotted in b versus CdSe core
size. All samples were excited
at 365 nm with a UV source.
For the 610-nm-emitting
QDs, this translates into a
Stokes shift of ~250 nm.
r = radius d, Comparison of
QD size to a MBP molecule.
555-nm-emitting CdSe/ZnS
core/shell QD, diameter ~60 Å,
surface-functionalized with
dihydrolipoic acid (red shell
~9–11 Å) has a diameter
~78–82 Å. The diagram
depicts the homogeneous
orientation MBP assumes
relative to the QD (Copyright
National Academy of Sciences
USA)53. MBP a midsize protein
(Mr ~ 44 kDa) has dimensions
of 30 × 40 × 65 Å (ref. 48).
nmat1390-print.indd 436 11/5/05 10:18:14 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 437
overcoat the native CdSe core with a layer of
wider-bandgap semiconducting material (for
example, ZnS and CdS)5,14,25. Fine-tuning this
synthetic scheme highlighted the importance of
the high-temperature solvent/ligand mixtures,
along with using less pyrophoric salt precursors
(CdO and Cd-acetate), for preparing reproducible
high-quality nanocrystals15,30.
QDs prepared using high-temperature routes
have no intrinsic aqueous solubility, thus phase-
transfer to aqueous solution requires surface
functionalization with hydrophilic ligands, either
through ‘cap exchange’, a process primarily driven
by mass action, or by encapsulating the original
nanocrystals in a thick heterofunctional organic
coating, driven mainly by hydrophobic absorption
onto the TOP/TOPO-capped QDs. These ligands
mediate both the colloid’s solubility and serve as
a point of chemical attachment for biomolecules.
Capping ligands serve another critical role in
insulating/passivating/protecting the QD surface
from deterioration in biological media. Synthesis
of new caps create an ever-increasing library
available for obtaining aqueous QD dispersions;
however, many were created for specialized uses
and thus have limited general applicability3,14,19,
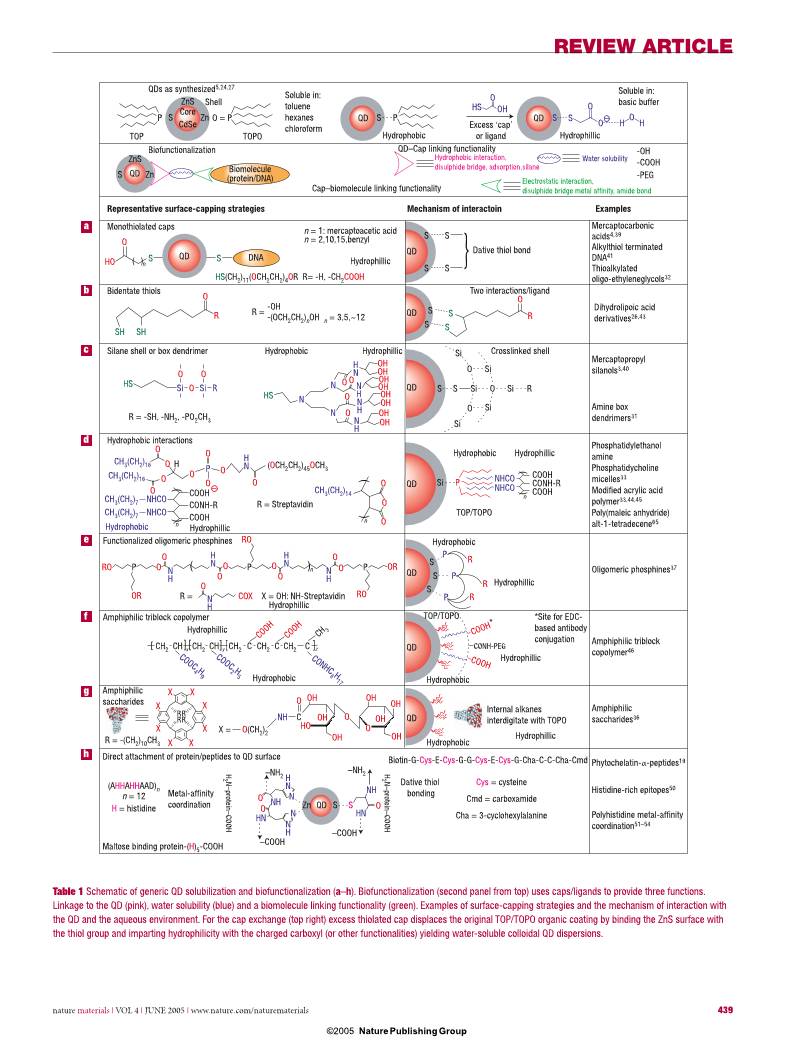
26,31–38. A representative list of caps and the QD-
dispersal strategies they use is provided in Table 1.
These strategies can be grouped into three major
routes. The fi rst uses ‘cap exchange’ and involves
the substitution of the native TOP/TOPO with
bifunctional ligands, each presenting a surface-
anchoring moiety to bind to the inorganic QD
surface (for example, thiol) and an opposing
hydrophilic end group (for example, hydroxyl,
carboxyl) to achieve water-compatibility. These
include an array of thiol and phosphine mono
and multidentate ligands (Table 1a,b)4,26,41–43. The
second strategy involves formation of polymerized
silica shells functionalized with polar groups,
which insulate the hydrophilic QD (Table 1c)3,40.
The third method preserves the native TOP/TOPO
on the QDs and uses variants of amphiphilic
‘diblock’ and ‘triblock’ copolymers and
phospholipids to tightly interleave/interdigitate
the alkylphosphine ligands through hydrophobic
attraction, whereas the hydrophilic outer
block permits aqueous dispersion and further
derivitization (Table 1d,f,g)33–36,44–46.
Is there a place for all these different capping
strategies? Probably, given that they address the
requirements of various potential applications.
But the advantages of each strategy have to be
carefully weighed against the drawbacks. For
example, compact mono-mercapto ligands
(Table 1a), although simple to synthesize, have
short shelf lives (<1 week) due to dynamic
thiol–ZnS interactions8. Substitution from mono
to dithiol dihydrolipoic acid ligands improves
long-term stability from ~1 week to 1–2 years,
suggesting that polydentate thiolated ligands
could be even more effective8,26,43. However, these
are carboxylic acids and almost all carboxy-
terminated ligands limit dispersion to basic
pHs8. Silica shells and polymer/phospholipid
encapsulation provides stability over a broader
pH range, but result in substantially larger
hydrophilic QDs. For example, phospholipid
and block copolymer coatings tend to increase
the diameter of CdSe–ZnS QDs from ~4–8 nm
before encapsulation to ~20–30 nm, a size that
although smaller than most mammalian cells can
still limit intracellular mobility and may preclude
Optical coding
Cellular and assay labelling
FRET donor Near-infrared imaging
Photoelectrochemistry
Microbiology-labels
visible400 700
ZnS CdSe
CdTe
PbS
PbSe/TeZnSe
CdS CdSe/Te CdHgTe alloys
Magnetic
FePt
Fe2O3
Co/SmCo5.2
UV < 250 2,500 > Infrared
λ (nm)
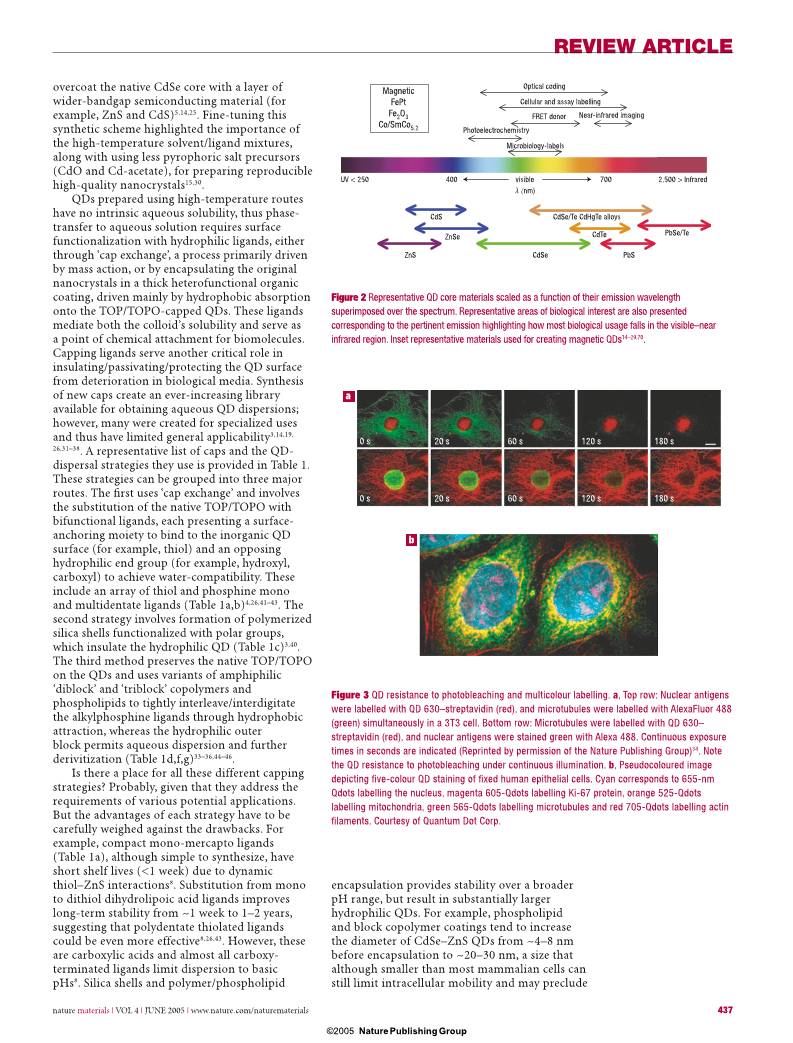
Figure 2 Representative QD core materials scaled as a function of their emission wavelength
superimposed over the spectrum. Representative areas of biological interest are also presented
corresponding to the pertinent emission highlighting how most biological usage falls in the visible–near
infrared region. Inset representative materials used for creating magnetic QDs14–29,70.
0 s 20 s 60 s 120 s 180 s
0 s 20 s 60 s 120 s 180 s
a
b
Figure 3 QD resistance to photobleaching and multicolour labelling. a, Top row: Nuclear antigens
were labelled with QD 630–streptavidin (red), and microtubules were labelled with AlexaFluor 488
(green) simultaneously in a 3T3 cell. Bottom row: Microtubules were labelled with QD 630–
streptavidin (red), and nuclear antigens were stained green with Alexa 488. Continuous exposure
times in seconds are indicated (Reprinted by permission of the Nature Publishing Group)34. Note
the QD resistance to photobleaching under continuous illumination. b, Pseudocoloured image
depicting fi ve-colour QD staining of fi xed human epithelial cells. Cyan corresponds to 655-nm
Qdots labelling the nucleus, magenta 605-Qdots labelling Ki-67 protein, orange 525-Qdots
labelling mitochondria, green 565-Qdots labelling microtubules and red 705-Qdots labelling actin
fi laments. Courtesy of Quantum Dot Corp.
nmat1390-print.indd 437 11/5/05 10:18:16 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
438 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
fluorescence resonance energy transfer (FRET)-
based investigations8. Lastly, an issue often
overlooked in biocompatible QD preparation is
monitoring of the ligand-exchange efficiency,
because performing effective cap exchange still
remains an art form. Clearly needed are systematic
analytical techniques evaluating the extent of cap
exchange beyond the final functional test19,46.
INORGANIC–BIOLOGICAL HYBRIDS
Inorganic–biological hybrids are made
by conjugating inorganic nanostructures
(nanoparticles, nanorods) with biomolecules
(proteins, DNAs) and the resulting conjugates
combine the properties of both materials,
that is, the spectroscopic characteristics of the
nanocrystal and the biomolecular function of
the surface-attached entities. Owing to its finite
size (comparable to or slightly larger than that
of many proteins, Fig. 1d) a single QD can be
conjugated to several proteins simultaneously.
The QD thus acts as a nanoscaffold for
attachment of several proteins, or other
biomolecules, creating a multifunctional
nanoparticle–biological hybrid. Our work has
shown that ~15–20 maltose binding proteins
(MBP, Mr ~ 44 kDa) can be attached
to each 6-nm-diameter QD (Fig. 1d)48. In
these assemblies, conjugate dimensions depend
on key parameters, such as surface cap used and
number and size of the biomolecules attached to
the surface. The function is dictated by the
nature of the biological moieties and their
conformation; features that have repercussions
on final performance. Proteins that do not
have their recognition site exposed away from the
QD surface may lose their ability to bind a target.
Conjugation schemes for attaching proteins to
QDs can be divided into three categories, each of
which has limitations. (i) Use of EDC, 1-ethyl-3-
(3-dimethylaminopropyl) carbodiimide,
condensation to react carboxy groups on the QD
surface to amines; (ii) direct binding to the QD
surface using thiolated peptides or polyhistidine
(HIS) residues; and (iii) adsorption or non-
covalent self-assembly using engineered proteins.
Conjugation using EDC condensation applied to
QDs capped with thiol-alkyl-COOH ligands often
produce intermediate aggregates due to poor QD
stability in neutral/acidic buffers. Using this same
chemistry for QDs encapsulated with polymeric
shells bearing COOH groups produces large
conjugates with poor control over the number
of biomolecules attached to a single nanocrystal.
Moreover, this chemistry is prone to crosslinking
and aggregating QDs, because the numerous
surface functional sites can bind/crosslink the
numerous protein target sites. Nonetheless,
this approach was used to prepare commercial
QD-streptavidin conjugates having ~20 proteins/
conjugate and relatively high quantum yield.
Streptavidin-coated QDs are used to attach
additional functionalities to the QDs (mostly
biotinylated antibodies), however they will bind
all biotinylated proteins indiscriminately allowing
only one targeted use.
Direct attachment of proteins/peptides to
the QD surface is based on two types of QD
surface–protein interaction; dative thiol-bonding
between QD surface sulphur atoms and cysteine
residues19,49 and metal-affi nity coordination
of HIS residues to the QD surface Zn atoms
(Table 1h)50,51. The Weiss group demonstrated the
former by using phytochelatin-related peptides
to cap CdSe/ZnS core/shell QDs, providing not
only surface passivation and water solubility,
but also a point of biochemical modifi cation
(Table 1h)19. Using peptides for both dispersion
and biofunctionalization may represent a new class
of rationally designed multifunctional biological
cap11,19. By using metal-affi nity coordination,
HIS-expressing proteins or peptides can be directly
attached to Zn on the QD-surface. This strong
interaction (Zn2+-HIS) has a dissociation constant,
KD, only slightly less than that measured for 6-
HIS to NTA-Ni2+ (10–13) but stronger than most
antibody bindings (10–6–10–9)51. Functional assays
with HIS-appended proteins indicate that control
can be exerted on the fi nal bioconjugate assembly
through the molar ratios of each participant added
before self-assembly48,52–54. This strategy allows
mixed protein surfaces, with the HIS affi nity for
other metals (Ni, Cu, Co, Fe, Mn) being relevant to
future materials51.
Engineering proteins to express positively
charged domains allows them to self-assemble
onto the surface of negatively charged QDs
through electrostatic assembly26. This approach
has proved useful for attaching a variety of
engineered proteins to QDs including MBP, for
purification over amylose resin, and Protein G,
which will bind the IgG portion of an antibody
thus acting as a linker55. Proteins can also be
non-specifically adsorbed to QDs56. Regardless
of the approach used in forming hybrid
conjugates, three important issues remain:
(i) the aforementioned lack of conjugation
strategies; (ii) the lack of homogeneity when
attaching proteins to QDs; and (iii) the inability
to finely control ratios of proteins/QD. The
lack of homogeneous attachment results in
heterogeneous protein orientation on the QD
surface, which may produce conjugates with
surface-attached proteins that are not optimally
functional. With antibodies, for example, many
may not be correctly oriented to bind their
intended target and this will manifest in low
avidity. It’s worth mentioning that success
in bioconjugation is intimately tied into
development of new caps, thus QD success in
biology will ultimately be driven by cap design.
EXISTING USES OF QUANTUM DOTS IN BIOLOGY
CELLULAR LABELLING
Cellular labelling is where QD use has made the
most progress and attracted the greatest interest.
Within the last two years, numerous reports have
nmat1390-print.indd 438 11/5/05 10:18:17 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 439
S
( (
( (
(
(
(
(
QDs as synthesized5,24,27
ZnS
Core
CdSe
Shell
P O = PS Zn
ZnS
S Zn
TOP TOPO
Soluble in:
toluene
hexanes
chloroform
QD
QD
QD
QD
QD
QD
QD
QD
S P QD S
Hydrophobic Hydrophillic
HS
O
O
O
O H H
OH
Excess ‘cap’
or ligand
–
–
Soluble in:
basic buffer
Biofunctionalization
QD
QD
Biomolecule
(protein/DNA)
Cap–biomolecule linking functionality
Hydrophobic interaction,
disulphide bridge, adsorption,silane
Electrostatic interaction,
disulphide bridge metal affinity, amide bond
QD–Cap linking functionality
Water solubility
-OH
-COOH
-PEG
DNA
O
HO S S
n
HS(CH2)11(OCH2CH2)4OR R= -H, -CH2COOH
n = 1: mercaptoacetic acid
n = 2,10,15,benzyl
Hydrophillic
S S
S S
S S
S S
Dative thiol bond
Mercaptocarbonic
acids4,39
Alkylthiol terminated
DNA41
Thioalkylated
oligo-ethyleneglycols32
Dihydrolipoic acid
derivatives26,43
Mercaptopropyl
silanols3,40
Amine box
dendrimers31
Bidentate thiols
Silane shell or box dendrimer
Monothiolated caps
SH SH
O
R R =
-OH
-(OCH2CH2)nOH n = 3,5,~12
O
R
Two interactions/ligand
HS Si Si RO
O O
R = -SH, -NH2, -PO2CH3
Hydrophobic
Hydrophobic
Hydrophobic
Hydrophillic
Hydrophillic
Hydrophillic
Hydrophillic
H
N
N
H
N
H
H
N
H
N
OH
OH
OH
OH
OH
OH
OH
OH
H
N
N
H
N
H
N
H
N
N
N
HS
OO
O
O
Crosslinked shellSi
Si Si
Si
Si
SiO
O R
O
Si P
S S
Hydrophobic interactions
CH3(CH2)16
CH3(CH2)16
CH3(CH2)14
CH3(CH2)7 NHCO
CH3(CH2)7 NHCO
O
O
O
O
O
O
O
O
O
O
O
O
H P (OCH2CH2)45OCH3
nn
R = Streptavidin
COOH
CONH-R
COOH
n
COOH
CONH-R
COOH
Hydrophobic
Hydrophobic
Hydrophobic
Hydrophobic
Hydrophillic
Hydrophillic
Hydrophillic
Hydrophillic
TOP/TOPO
TOP/TOPO
Phosphatidylethanol
amine
Phosphatidycholine
micelles33
Modified acrylic acid
polymer33,44,45
Poly(maleic anhydride)
alt-1-tetradecene65
Oligomeric phosphines37
S
S
S
P
P
P
R
R
R
RO P P P
RO
RO
O
O O
O
O
O
O OR
O
O
OR
( )n
N
H
R = COX X = OH: NH-Streptavidin
Functionalized oligomeric phosphines
Amphiphilic triblock copolymer *Site for EDC-
based antibody
conjugation
COOH
COOH*
CONH-PEG
Amphiphilic triblock
copolymer46
X
X X
X
XX
X X
R
RR
R
Amphiphilic
saccharides
NH C
O OH
OH
OH
OH
OH
OH
OH
HO
O
OX = O(CH2)2
Internal alkanes
interdigitate with TOPO
Amphiphilic
saccharides36
O
N
H
H
N
O
O
NH
NH
HNHN
N
N
Zn QD S S
–COOH
–COOH
–NH2–NH2
Direct attachment of protein/peptides to QD surface
(AHHAHHAAD)n
n = 12
H = histidine
Metal-affinity
coordination
Maltose binding protein-(H)5-COOH
Biotin-G-Cys-E-Cys-G-G-Cys-E-Cys-G-Cha-C-C-Cha-Cmd
H
2 N–protein–COOH
H
2 N–protein–COOH
Dative thiol
bonding
Cys = cysteine
Cmd = carboxamide
Cha = 3-cyclohexylalanine
Phytochelatin-α-peptides19
Histidine-rich epitopes50
Polyhistidine metal-affinity
coordination51–54
CH2 CCH2CH2 CH2 CH2CH CH C C[ [ [] ] ]
CH 3
CONHC
8 H
17
COOC
4 H
9
COOC
2 H
5
COOH
COOH
R = -(CH2)10CH3
Representative surface-capping strategies Mechanism of interactoin Examples
NHCO
NHCO
zyx
a
b
c
d
e
f
g
h
Table 1 Schematic of generic QD solubilization and biofunctionalization (a–h). Biofunctionalization (second panel from top) uses caps/ligands to provide three functions.
Linkage to the QD (pink), water solubility (blue) and a biomolecule linking functionality (green). Examples of surface-capping strategies and the mechanism of interaction with
the QD and the aqueous environment. For the cap exchange (top right) excess thiolated cap displaces the original TOP/TOPO organic coating by binding the ZnS surface with
the thiol group and imparting hydrophilicity with the charged carboxyl (or other functionalities) yielding water-soluble colloidal QD dispersions.
nmat1390-print.indd 439 11/5/05 10:18:18 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
440 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
appeared describing the ability of one or more
‘colour/size’ of biofunctionalized QDs to label
cells. Many of these reports show that QD labelling
permits extended visualization of cells under
continuous illumination as well as multicolour
imaging, highlighting the advantages offered by
these fl uorophores (Table 2, Fig. 3)34,56–60. A clear
differentiation can be made between labelling of live
or ‘fi xed’ cells (dead, with chemically crosslinked
components to maintain cellular architecture).
Fixed cells can be treated ‘harshly’ to facilitate
entry of the QD reagent by chemically creating
pores. For labelling live cells, the process must be
handled judiciously to maintain cellular viability.
The major hurdle is entry of the relatively large
QDs into the cell across the cellular membranes’
lipid bilayer. Strategies to accomplish this include
non-specifi c uptake by endocytosis, where QDs
often end up in endocytic compartments (Table 2);
direct microinjection of nanolitre volumes, which
is tedious and limits the number of cells labelled;
electroporation, which uses charge to physically
deliver QDs through the membrane and mediated/
targeted uptake61–63. Mediated uptake uses reagents
such as Lipofectamine 2000, which encapsulates
QDs within lipid vesicles to facilitate entry into the
cell63. Targeted uptake exploits the cells’ propensity
to recognize and internalize QDs labelled with
specifi c peptides (for example, HIV-derived TAT
peptide) and even deliver them to specifi c cellular
compartments such as the nucleus61. Comparison
of several of these methods showed that the
Table 2 Representative cellular components and proteins that have been labelled with QDs. *Indicates that labelling was
performed in live cells.
Cellular component/protein Function
Nucleus/nuclear proteins* Internal organelle containing the genetic information (chromosomal
DNA), enclosed by a membrane containing pores that mediate
transport in and out34,61,62. See Fig. 3.
Mitochondria* Organelle providing essential energy-delivery processes, contains its
own genome62. See Fig. 3.
Microtubules Cytoskeleton protein that maintains cellular structure and reforms
during movement34. See Fig. 3.
Actin filaments Cytoskeleton protein that maintains cellular structure, helps localize
other organelles and reforms during movement34. See Fig. 3.
Endocytic compartments* Vesicles that form on cell surfaces and mediate both specific and non-
specific internalization of extracellular molecules56,58.
Mortalin Member of Heat Shock 70 Protein Family. Differential staining pattern
in normal and precancerous cells59.
Cytokeratin Cytoskeleton protein that is overexpressed and differentially stained in
many skin cancer cells57.
Cellular membrane proteins and receptors This membrane is a lipid bilayer that encloses the cytoplasm (cellular
fluid environment) and contains the membrane spanning channels and
receptors that allow the cell to communicate with its environment.
Serotonin transport proteins* Cell surface transporter of serotonin and related neurotransmitters69.
Prostate-specific membrane antigen* Protein expressed on surface of normal and cancerous prostate cells46.
Her2* Breast cancer marker protein overexpressed on the surface of some
breast cancer cells34.
Glycine receptors* Main inhibitory neurotransmitter receptor on surface of spinal nerve
cells66.
erbB/HER* Cellular membrane spanning receptor that mediates cellular response
to growth factors. Once activated, the receptors undergo endocytosis
and recycling or degradation. Overexpressed in many cancers67.
p-glycoprotein* Multidrug transporter that spans the membrane and is a mediator of
multidrug resistance in cancer cells57,58.
nmat1390-print.indd 440 11/5/05 10:18:18 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 441
mediator Lipofectamine 2000 had the highest
delivery effi ciency, however, the QDs were delivered
in aggregates62,63. Others combined techniques
by electroporating QDs covered with a nuclear
localization sequence into cells and then monitored
their translocation into the nuclear compartment61.
Peptide-targeted uptake looks to be the most specifi c
for delivering dispersed QDs into cells, although
it may be limited to cells expressing appropriate
receptors. Once delivered inside the cytoplasm of
cells, dispersion of the QDs depends strongly on
their surface coating and pH stability. QDs capped
with COOH-terminated groups often aggregate
shortly after introduction, due to their poor stability
in acidic conditions, whereas protein-coated QDs are
more dispersed in the cytoplasm.
To demonstrate multicolour labelling of live
cells, endocytic uptake and selective labelling of
cell surface proteins were used with antibody-
conjugated QDs, and then QD emission was
followed for over a week as cellular development
was monitored, showing that cells tolerate QDs for
extended periods of time33,58,65. By using antibody-
linked QDs, both the Her2 breast cancer marker
and specifi c intracellular proteins were labelled in
both live and fi xed cancer cells, demonstrating that
QD-probes can cross into cells and specifi cally bind
their intended intracellular targets34. QDs can also
be useful markers for tracking cellular movement,
differentiation and fate33,58,65. Cancer cells seeded
on top of QDs engulf them and leave behind
a fl uorescence-free phagokinetic trail65. Using
several cell lines, the size and shape of trails were
shown to correlate well with potential invasiveness,
providing a novel assay for this property65. Micelle-
stabilized QDs injected into Xenopus embryos
demonstrated cell lineage-tracing during the
complex developmental process spanning the
embryo–tadpole stage and equal transfer of QDs
from mother to daughter cells, again showing long-
term cellular compatibility even with >109 QDs per
cell33. By exploiting QD photobleaching resistance,
previously onerous cellular physiology questions
have been investigated on the single-molecule level.
These include tracking the diffusion dynamics of
individual glycine receptors in neuronal cells over
long periods of time (min), which provided unique
information on single-protein movement, and
interestingly, the authors used blinking to identify
single QDs and differentiate them from aggregates66.
Additionally, the specifi c erbB/HER cellular
receptor-mediated fusion and internalization
process has been observed in single cells67. These
publications mark the transition from ‘proof-of-
concept’ to useful tool as these complex cellular
processes could not be tracked continuously with
photolabile organic fl uorophores.
IN VIVO AND DEEP TISSUE IMAGING
Imaging tissue with far red/near infrared
excitation overcomes some problems due to
indigenous tissue autofluorescence, and QD
spectroscopic properties can be exploited here
to achieve somewhat deeper penetration than
the available near-infrared dyes68–70. This was
demonstrated by synthesizing near-infrared-
emitting QDs (840–860 nm) and applying them to
sentinel lymph-node mapping in cancer surgery of
animals70. Using only 5 mW cm–2 excitation, they
imaged lymph nodes 1 cm deep in tissue, where
lymphatic vessels were clearly visualized draining
QD solutions into the sentinel nodes (Fig. 4)70. As
an added bonus, the location of QD accumulation
in the excised nodes may be the most likely
place for the pathologist to find metastatic cells.
Interestingly, QDs have been demonstrated to
remain fluorescent in tissues in vivo for four
months45. In addition to the properties described
above, QDs have large two-photon cross-sectional
efficiency with a two-photon fluorescence process
100–1,000× that of organic dyes. This makes them
suitable for in vivo deep-tissue imaging using
two-photon excitation at low intensities63,71. Using
this technique to excite green emitting QDs in the
near infrared allowed imaging of mice capillaries
hundreds of micrometres deep and subcellular
resolution of mouse brain68,71.
QDs may also be useful for tracking cancer cells
in vivo during metastasis46,60,63. A multifunctional
QD probe has been developed46 (Table If) linked
with tumour-targeting antibodies. In vivo studies in
mice expressing human cancer showed that these
QD probes accumulated at the tumour sites. Using a
slightly different approach, tumour cells were labelled
with QDs, injected into mice and then tracked with
multiphoton microscopy as they invaded lung tissue63.
In both studies, spectral imaging and autofl uorescent
subtraction allowed multicolour in vivo visualization
of cells and tissues46,63. The ability to track cells in vivo
without continuously sacrifi cing animals represents
a substantial improvement over current techniques.
1 cm
Colour video Near-infrared fluorescence Colour/near-infrared merge
Au
to
flu
or
es
ce
nc
e
30
s
ec
p
os
t-
in
je
ct
io
n
4
m
in
po
st
-in
je
ct
io
n
Im
ag
e-
gu
id
ed
re
se
ct
io
n
Figure 4 Near-infrared QD
imaging in vivo. Images of
the surgical fi eld in a pig
injected intradermally with
400 pmol of near-infrared
QDs in the right groin. Four
time points are shown from
top to bottom: before injection
(autofl uorescence), 30 s after
injection, 4 min after injection
and during image-guided
resection. For each time point,
colour video (left), near-infrared
fl uorescence (middle) and
colour-near infrared merge
(right) images are shown. Note
the lymphatic vessel draining
to the sentinel node from
the injection site. (Reprinted
by permission of the Nature
Publishing Group)70.
nmat1390-print.indd 441 11/5/05 10:18:19 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
442 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
Although QDs are clearly superior to dyes for these
purposes, the question of whether the large QD probe
mirrors true in vivo physiology remains unanswered.
There are many open questions about the
toxicity of inorganic QDs containing Cd, Se, Zn,
Te, Hg and Pb72,73. These can be potent toxins,
neurotoxins and teratogens depending on dosage
and complexation, accumulating in and damaging
the liver and nervous system. In small dosages,
the metals are bound by metallothionein proteins
and may be excreted slowly or sequestered in
vivo in adipose and other tissues64,72. Although
cellular studies have followed QD exposure over
time33,34,58,60,63, to date there have been no long-
term animal studies assessing QD toxicology. CdSe
QD toxicity has been examined using a hepatocyte
culture model and it was found that exposure of
core CdSe to an oxidative environment caused
decomposition and desorption of Cd ions,
whereas adding a shell of 1–2 monolayers of ZnS
reduced oxidation, but did not fully eliminate
cytotoxicity induced by 8 h of photooxidation64.
A rather thick ZnS-overcoating (4–6 monolayers)
in combination with efficient surface capping/
coating can substantially reduce desorption of
core ions; nonetheless, extreme radiation can still
cause desorption of the core ions64. Additionally, it
has been reported that QDs could damage DNA74
and that QD surface coatings effect cytotoxicity75.
Given the broad interest in QDs, these results are
fuelling a strong debate on this issue64,74,75. The
fact that some long-term in vivo studies have not
found evidence of toxicity is promising33,45,58,63,
but still not a ‘blanket’ endorsement. For in vivo
imaging, if the metals are safely contained, the
issue will become one of metabolically clearing
the nanoparticle, however, hardly anything is
known about how these particles will be cleared
by the body72,73. Given the myriad materials,
preparations and coatings, it is debatable as to
whether the definitive comprehensive study will
ever be realized.
QD ASSAY LABELLING
QD assay labelling uses QDs for in vitro
assay detection of DNA, proteins and other
biomolecules. DNA-coated QDs have been shown
as sensitive and specific DNA labels for in situ
hybridizations47, as probes for human metaphase
chromosomes76, and in single-nucleotide
polymorphism and multi-allele DNA detection77.
Conversely, DNA linked to QD surfaces has been
used to code and sort the nanocrystals78. These
results demonstrate that DNA-conjugated QDs
specifically bind their complements both in
fixed cells and in vitro. For proteins, our group
uses a self-assembled electrostatic protein QD-
functionalization strategy to create QD-based
fluoro-immunoassay reagents. Using antibodies
conjugated to QDs by an adaptor protein, we have
carried out multiple demonstrations of analyte
detection in numerous immunoassays including
a four-colour multiplex toxin analysis42,79. For
bioassays, the ability to excite QDs at almost any
wavelength below the band edge combined with
high photobleaching resistance and ‘multiplexing’
capabilities highlights the unique combination of
spectral properties that make QD-fluorophores of
interest to biologist.
QDS AND FRET
As FRET is sensitive to molecular rearrangements
on the 1–10 m range (a scale correlating to the size
of biological macromolecules), researchers have
long used this photophysical process to monitor
intracellular interactions and binding events1.
Reports of QDs as FRET donors in a biological
context appeared quickly39,80,81; however, the full
potential has only been demonstrated recently. By
self-assembling acceptor dye-labelled proteins onto
QD donor surfaces, two unique advantages over
organic fl uorophores for FRET became apparent:
QD donor emission could be size-tuned to improve
spectral overlap with a particular acceptor dye, and
having several acceptor dyes interact with a single
QD-donor substantially improved FRET effi ciency
(Fig. 5a,b)52. The scenario in Fig. 5b demonstrates
the latter using a 30 Å radius QD with a dye-
labelled protein attached to the QD surface and
the dye located at 70 Å from the core. Assuming
a Förster distance (R0) for this QD donor–dye
acceptor pair of 56 Å, then the FRET effi ciency
for a single donor–acceptor pair would be 22%.
By increasing the number of acceptors to fi ve, the
effi ciency more than doubles to ~58%48,52.
QDs also function as effective protein
nanoscaffolds and exciton donors for prototype
self-assembled FRET nanosensors targeting the
nutrient maltose by using MBP48. QDs could
even drive biosensors through a two-step FRET
mechanism overcoming inherent donor–acceptor
distance limitations, as schematically depicted in
Fig. 5c, where each 530-nm QD is surrounded by
~10 MBPs (each mono-labelled with Cy3, one
shown for clarity)48. β-cyclodextrin-Cy3.5 (β-
CD-Cy3.5) specifi cally bound in the MBP central
binding pocket completes the QD-10MBP-Cy3-β-
CD-Cy3.5 sensor complex. Excitation of the QD
results in FRET excitation of the MBP-Cy3, which
in turn FRET-excites the β-CD-Cy3.5 overcoming
the low direct QD-Cy3.5 FRET. Added maltose
displaces β-CD-Cy3.5 leading to increased Cy3
emission. Using a set of site-specifi cally dye-
labelled MBPs and a FRET strategy analogous to a
nanoscale global positioning system determination,
the QD-MBP structure was modelled and the
results indicate that MBP assumes a homogeneous
orientation once assembled on the QD surface
(Fig. 1d)53. This result suggests that it’s possible
to self-assemble QD–protein conjugates with the
proteins homogeneously oriented; an important
fi nding in light of previous concerns about forming
hybrid structures.
Paradoxically, the excellent QD donor properties
(long fl uorescent lifetime, broad absorption and high
extinction coeffi cient) may almost preclude their role
as FRET acceptors for organic dyes82. The larger size
of ‘redder’ CdSe and other QDs (emission >600 nm,
nmat1390-print.indd 442 11/5/05 10:18:20 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 443
Many biological applications of QDs have just begun to be explored. Using combinations of QD emissions
to create multicolour optical barcodes is promising, as estimates indicate that with just six colours in 10
different intensities, a library of ~1 million entries could be encoded89. However, this remains ‘proof-
of-concept’ with preliminary reports showing that polystyrene beads optically encoded with QDs and
targeting DNA can be hybridized and detected at the single bead level89. The Willner group have been
investigating QD photoelectrochemistry using DNA-driven QD arrays to harness photogenerated
currents in the pursuit of novel optoelectronic devices90. In an elegant display, CdS QDs were bound in
the central pocket of a GroEL chaperonin protein complex, which released this cargo on the addition of
ATP (Fig. B1a,b)91. GroEL is a cylindrical chaperonin protein complex (Mr ~840 kDa, 4.5 nm cavity) that
functions in refolding denatured proteins in vivo91. The schematic representation shows the formation
of GroEL–CdS nanoparticle complexes through inclusion of CdS nanoparticles into the cylindrical
cavity of GroEL, and its ATP-triggered guest release along with transmission electron micrographs of
these chaperonin–CdS nanoparticle complexes. The models on the sides of the images are schematic
representations of the end-view of the complexes (Reprinted by permission of the Nature Publishing
Group)91. This demonstration bodes well for developing a biosensor or intracellular QD sequestering and
delivery mechanism based on controlled QD release in response to a target analyte.
The drive to combine the ability to capture, separate and visualize cells (or proteins) within one reagent
is driving interest in biomagnetic QDs. One approach used antibiotic-coated FePt particles to capture and
identify bacteria at ultralow concentrations, whereas another used nitriloacetic-acid-coated FePt particles to
effi ciently capture HIS-appended proteins17,21. In an attempt at hybrid magnetic–luminescent QD complexes,
polymer-coated Fe2O3 cores overcoated with a CdSe-ZnS QD shell and functionalized with antibodies were
used to magnetically capture breast cancer cells, which were then viewed fl uorescently92. A unique bis-
functional CdS-FePt luminescent–magnetic dimeric QD particle has been synthesized, however, its biological
function remains to be explored18. See Fig. B1c for a schematic and high-resolution TEM of the QD-
magnetic CdS-FePt nanoparticles. QDs are also promising tools for visualizing in vitro protein movements,
such as sliding of actin fi laments93,94, as strain-specifi c microbiological labels95 and for enzymatic monitoring
of DNA replication96. QDs have even been tested as contrasting agents for fi ngerprint dusting97.
Box 1 Developing technologies
50 nm
5 nm
CdS
nanoparticle
ATP
Mg2+ +K+
Coagulation
Release
GroEL/CdS nanoparticle complex
a
b c CdS-FePt
Figure B1 Novel QD bio-
applications and materials.
a, GroEL is a cylindrical
chaperonin protein complex
(MW ~840 kDa, 4.5 nm
cavity) that functions
in refolding denatured
proteins in vivo91. Schematic
representation of the
formation of GroEL–CdS
nanoparticle complexes by
inclusion of CdS nanoparticles
into the cylindrical cavity
of GroEL, and its ATP-
triggered guest release.
b, Transmission electron
micrographs of chaperonin–
CdS nanoparticle complexes.
The models on the sides of
the images are schematic
representations of the
end-view of the complexes
(Reprinted by permission
of the Nature Publishing
Group)91. c, Schematic
and high-resolution TEM
of QD-magnetic CdS–FePt
nanoparticles (Reprinted with
permission of the American
Chemical Society)18.
nmat1390-print.indd 443 11/5/05 10:18:21 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
444 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
510 510 510 510570 570 570 570
0.5 ns 2.5 ns 4.5 ns 6.5 ns
nR0
6
nR0
6 + r 6
Eff =
R0 = 56 Å
r (Å) n Eff (%)
70 21.6
58.05
1
70
Protein
Dye
70 Å
30
Å
QD
R0 = 56 Å
2R0
1.0
0.5
0
Ef
fic
ie
nc
y
56 70
Distance (Å)
530
QD
530
QD
Fret 1 Fret 1
Fret 2Excitation Excitation
Cy3 Cy3
Cy3.5 Cy
3.
5
Emission
Emission
Maltose
MBP
MBP
β-Cyclodextrin-Cy3.5
Initiation of apoptosis
DNA degradation
Cancer cell
Antibody
Quantum dot
Quantum dot
Energy transfer
Classical
photosensitizer
hν
hν
hν
3O2
3O2
3O2
1O2
1O2
Cd2+
Cd2+
ROS
ROS
a
b
c
d
5HIS
5HIS
Figure 5 Properties of QDs as FRET donors. a, Images displaying FRET between a 510-nm-
emitting QD donor and MBP-Cy3 acceptor immediately after a short excitation pulse as recorded
by the CCD camera at 2-ns intervals. Panel 1: 510-nm QD donor only, Panel 2: MBP-Cy3
acceptor only, Panel 3: 510-nm QD donor with MBP-Cy3 acceptor self-assembled on the QD
surface. (Reprinted by permission of the American Chemical Society)52. b, Demonstration of
improved FRET efficiency (Eff) derived from arraying multiple acceptor dyes around a single
QD donor acting as a protein scaffold. c, Schematic of a 530QD-MBP-Cy3-β-CD-Cy3.5 maltose
sensing assembly. (Reprinted by permission of the Nature Publishing Group)48. d, Schematic
of how QD photosensitizers functionalized with cancer cell-specific antibodies would bind and
specifically kill a cancer cell in vivo. The antibodies direct the QDs to bind only to cancer cells
and QDs then harvest either UV or infrared energy (by two-photon excitation) and use this to
generate reactive oxygen species, ROS, which initiate apoptosis or programmed cell death
(Figure kindly provided by R. Bakalova)84.
diameter >8 nm) may also preclude FRET as the
Förster distance (R0, donor/acceptor distance for 50%
energy transfer) may fall within the core–shell radius,
suggesting an upper limit on QD size for FRET52,53.
Beyond sensors, QD-FRET may have potential
medical usage as photosensitizers in photodynamic
medical therapy (see Fig. 5d)83,84.
Other potential QD technologies are outlined
in Box 1.
FUTURE OUTLOOK
Cellular labelling will continue to see substantial
progress. Studies using ‘extensive’ multiplexing
(6–10 colours) will focus on elucidating complex
cellular processes by exploiting QD photobleaching
resistance and multicolour resolution. Cellular
events will be studied on the single biomolecule
level with QDs, although intermittency may become
an issue for these types of experiments. Work on
near-infrared dots as tissue probes and contrast
agents will continue almost exclusively in animal
models due to unsettled issues of toxicity, which
will remain a hot issue without a defi nitive near-
term answer. Progress in optical bar-coding will
encompass some combinatorial chemistry synthesis
scheme with the QDs functioning as barcodes for the
synthetic products. Application of QD bar-coding
to high-throughput or parallel assay formats, such
as gene-expression monitoring or drug-discovery
assays, can be anticipated. The newly available
commercial Qbead and Mosaic Gene Expression
technology will stimulate use, however, the cost
of the dedicated support equipment needed for
decoding may limit broader applications85. An
interesting derivative of QD-barcodes may include
magnetic properties for capture. The bifunctional
magnetic/luminescent QDs are intriguing and
potentially make an ideal reagent for capturing
and visualizing low-copy-number biomolecules.
Biofunctionalized paramagnetic nanocrystals
targeted to particular in vivo sites, such as tumours,
may make novel magnetic resonance imaging agents.
Using bifunctional QDs for concurrent magnetic
resonance imaging and deep-tissue imaging may
radically alter in vivo imaging86. FRET-based QD-
biosensors will migrate intracellularly to monitor
physiological processes in real-time. Interestingly, the
QD multiplexing potential remains almost untapped
and so bioassays and studies using more than fi ve
colours simultaneously can be expected.
Another intriguing area involves QD ‘blinking’
or intermittency. Epitaxial QDs do not blink. Recent
reports suggest that thiol-reducing agents may
suppress colloidal QD blinking even in biological
buffers, further suggesting that this process may be
modulated12,13,87. Understanding of QD blinking
processes will slowly be elucidated, and harnessing
this phenomenon may create biosensors that truly
switch off and on. Use of novel QD rod materials with
polarized emission may be of benefi t to fl uorescence
polarization assays88. We can also envisage complex
QD-bioarrays that incorporate biological functions to
be fully integrated into nanodevices for light/energy
harvesting, biosensing or molecular electronics.
nmat1390-print.indd 444 11/5/05 10:18:21 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials 445
Will QDs replace fl uorescent dyes? No — rather
they will complement dye defi ciencies in particular
applications such as in vivo imaging. Moreover,
adapting QDs for biological use will teach us critical
lessons about creating future inorganic–biological
hybrids with direct relevance to many other materials.
QDs have far from exhausted their biological
potential. The shift to enabling everyday research is
now beginning, mostly driven by cellular labelling.
Other areas, such as medical imaging, FRET
biosensing, assay labelling and optical barcoding are
likely to follow suit.
Note added in proof: While this manuscript was
in preparation, ref. 99 was published. It provides an
excellent overview of QD usage for in vivo imaging
and diagnostics.
doi:10.1038/nmat1390
References
1. Miyawaki, A. Visualization of the spatial and temporal dynamics of
intracellular signaling. Dev. Cell 4, 295–305 (2003).
2. Schrock, E. et al. Multicolor spectral karyotyping of human chromosomes.
Science 273, 494–497 (1996).
3. Bruchez, M., Jr, Moronne, M., Gin, P., Weiss, S. & Alivisatos, A. P.
Semiconductor nanocrystals as fl uorescent biological labels. Science 281,
2013–2016 (1998).
4. Chan, W. C. W. & Nie, S. Quantum dot bioconjugates for ultrasensitive
nonisotopic detection. Science 281, 2016–2018 (1998).
5. Dabbousi, B. O. et al. (CdSe)ZnS core-shell quantum dots: synthesis and
optical and structural characterization of a size series of highly luminescent
materials. J. Phys. Chem. B 101, 9463–9475 (1997).
6. Leatherdale, C. A., Woo, W. K., Mikulec, F. V. & Bawendi, M. G. On the
absorption cross section of CdSe nanocrystal quantum dots. J. Phys. Chem. B
106, 7619–7622 (2002).
7. Murphy, C. J. Optical sensing with quantum dots. Anal. Chem. 74, 520A–526A
(2002).
8. Parak, W. J. et al. Biological applications of colloidal nanocrystals. Nanotech.
14, R15–R27 (2003).
9. Niemeyer, C. M. Nanoparticles, proteins, and nucleic acids: biotechnology
meets materials science. Angew. Chem. Int. Edn Eng. 40, 4128–4158 (2001).
10. Alivisatos, P. The use of nanocrystals in biological detection. Nature Biotechnol.
22, 47–52 (2004).
11. Mattoussi, H., Kuno, M. K., Goldman, E. R., Anderson, G. P. & Mauro, J. M. in
Optical Biosensors: Present and Future (eds Ligler, F. S. & Rowe C. A.) 537–569
(Elsevier, Amsterdam, Netherlands, 2002).
12. Nirmal, M. et al. Fluorescence intermittency in single cadmium selenide
nanocrystals. Nature 383, 802–806 (1996).
13. Efros, A. L. & Rosen, M. Random telegraph signal in the photoluminescence
intensity of a single quantum dot. Phys. Rev. Lett. 78, 1110–1113 (1996).
14. Hines, M. A. & Guyot-Sionnest, P. Synthesis and characterization of strongly
luminescing ZnS-Capped CdSe nanocrystals. J. Phys. Chem. 100, 468–471 (1996).
15. Peng, Z. A. & Peng, X. Formation of high-quality CdTe, CdSe, and CdS
nanocrystals using CdO as precursor. J. Am. Chem. Soc. 123, 183–184 (2001).
16. Bailey, R. E. & Nie, S. Alloyed semiconductor quantum dots: tuning the
optical properties without changing the particle size. J. Am. Chem. Soc. 125,
7100–7106 (2003).
17. Gu, H., Ho, P.-L., Tsang, K. W. T., Wang, L. & Xu, B. Using Biofunctional
magnetic nanoparticles to capture vancomycin-resistant enterococci and
other gram-positive bacteria at ultralow concentration. J. Am. Chem. Soc. 125,
15702–15703 (2003).
18. Gu, H., Zheng, R., Zhang, X. & Xu, B. Facile one-pot synthesis of bifunctional
heterodimers of nanoparticles: A conjugate of quantum dot and magnetic
nanoparticles. J. Am. Chem. Soc. 126, 5664–5665 (2004).
19. Pinaud, F., King, D., Moore, H.-P. & Weiss, S. Bioactivation and cell targeting
of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides.
J. Am. Chem. Soc. 126, 6115–6123 (2004).
20. Tsay, J. M., Pfl ughoefft, M., Bentolila, L. A. & Weiss, S. Hybrid approach to the
synthesis of highly luminescent CdTe/ZnS and CdHgTe/ZnS nanocrystals. J.
Am. Chem. Soc. 126, 1926–1927 (2004).
21. Xu, C. J. et al. Nitrilotriacetic acid-modifi ed magnetic nanoparticles as a general
agent to bind histidine-tagged proteins. J. Am. Chem. Soc. 126, 3392–3393 (2004).
22. Murray, C. B., Kagan, C. R. & Bawendi, M. G. Synthesis and characterization
of monodisperse nanocrystals and close-packed nanocrystal assemblies. Ann.
Rev. Mater. Sci. 30, 545–610 (2000).
23. Xu, C. J. et al. Dopamine as a robust anchor to immobilize functional
molecules on the iron oxide shell of magnetic nanoparticles. J. Am. Chem. Soc.
126, 5664–5665 (2004).
24. Murray, C. B., Norris, D. J. & Bawendi, M. G. Synthesis and characterization
of nearly monodisperse CdE (E = sulfur, selenium, tellurium) semiconductor
nanocrystallites. J. Am. Chem. Soc. 115, 8706–8715 (1993).
25. Peng, X., Schlamp, M. C., Kadavanich, A. V. & Alivisatos, A. P. Epitaxial growth
of highly luminescent CdSe/CdS core/shell nanocrystals with photostability
and electronic accessibility. J. Am. Chem. Soc. 119, 7019–7029 (1997).
26. Mattoussi, H. et al. Self-assembly of CdSe-ZnS quantum dot bioconjugates using
an engineered recombinant protein. J. Am. Chem. Soc. 122, 12142–12150 (2000).
27. Hines, M. A. & Guyot-Sionnest, P. Bright UV-blue luminescent colloidal ZnSe
nanocrystals. J. Phys. Chem. B 102, 3655–3657 (1998).
28. Suyver, J. F., Wuister, S. F., Kelly, J. J. & Meijerink, A. Synthesis and
photoluminescence nanocrystalline ZnS:Mn2+. Nano Lett. 1, 429–433 (2001).
29. Artmeyev, M. V., Gaponenko, S. V., Germanenko, I. N. & Kapitonov, A. M.
Irreversible photochemical spectral hole burning in quantum-sized CdS
nanocrystals embedded in a polymeric fi lm. Chem. Phys. Lett. 243, 450–455
(1995).
30. Reiss, P., Bleuse, J. & Pron, A. Highly luminescent CdSe/ZnSe core/shell
nanocrystals of low size dispersion. Nano Lett. 2, 781–784 (2002).
31. Guo, W., Li, J. J., Wang, Y. A. & Peng, X. Conjugation chemistry and
bioapplications of semiconductor box nanocrystals prepared via dendrimer
bridging. Chem. Mater. 15, 3125–3133 (2003).
32. Hong, R. et al. Control of protein structure and function through surface
recognition by tailored nanoparticle scaffolds. J. Am. Chem. Soc. 126, 739–743
(2004).
33. Dubertret, B. et al. In vivo imaging of quantum dots encapsulated in
phospholipids micelles. Science 298, 1759–1762 (2002).
34. Wu, X. et al. Immunofl uorescent labeling of cancer marker Her2 and other
cellular targets with semiconductor quantum dots. Nature Biotechnol. 21,
41–46 (2003).
35. Pellegrino, T. et al. Hydrophobic nanocrystals coated with an amphiphilic
polymer shell: A general route to water soluble nanocrystals. Nano Lett. 4,
703–707 (2004).
36. Osaki, F., Kanamori, T., Sando, S., Sera, T. & Aoyama, Y. A quantum dot
conjugated sugar ball and its cellular uptake on the size effects of endocytosis
in the subviral region. J. Am. Chem. Soc. 126, 6520–6521 (2004).
37. Kim, S. & Bawendi, M. G. Oligomeric ligands for luminescent and stable
nanocrystal quantum dots. J. Am. Chem. Soc. 125, 14652–14653 (2003).
38. Wang, X.-S. et al. Surface passivation of luminescent colloidal quantum dots
with poly(dimethylaminoethyl methacrylate) through a ligand exchange
process. J. Am. Chem. Soc. 126, 7784–7785 (2004).
39. Willard, D. M., Carillo, L. L., Jung, J. & Van Orden, A. CdSe-ZnS quantum dots
as resonance energy transfer donors in a model protein-protein binding assay.
Nano Lett. 1, 469–474 (2001).
40. Gerion, D. et al. Synthesis and properties of biocompatible water-soluble
silicacoated CdSe/ZnS semiconductor quantum dots. J. Phys. Chem. B 105,
8861–8871 (2001).
41. Mitchell, G. P., Mirkin, C. A. & Letsinger, R. L. Programmed assembly of DNA
functionalized quantum dots. J. Am. Chem. Soc. 121, 8122–8123 (1999).
42. Goldman, E. R. et al. Avidin: a natural bridge for quantum dot-antibody
conjugates. J. Am. Chem. Soc. 124, 6378–6382 (2002).
43. Uyeda, H. T., Medintz, I, L., Jaiswal, J. K., Simon, S. M. & Mattoussi H.
Synthesis of compact multidentate ligands to prepare stable hydrophilic
quantum dot fl uorophores. J. Am. Chem. Soc. 127,3870–3878 (2005).
44. Mattheakis, L. C. et al. Optical coding of mammalian cells using
semiconductor quantum dots. Anal. Biochem. 327, 200–208 (2004).
45. Ballou, B., Lagerholm, B. C., Ernst, L. A., Bruchez, M. P. & Waggoner, A. S.
Noninvasive imaging of quantum dots in mice. Bioconj. Chem. 15, 79–86
(2004).
46. Gao, X., Cui, Y., Levenson, R. M., Chung, L. W. K. & Nie, S. In vivo cancer
targeting and imaging with semiconductor quantum dots. Nature Biotechnol.
22, 969–976 (2004).
47. Pathak, S., Choi, S. K., Arnheim, N. & Thompson, M. E. Hydroxylated
quantum dots as luminescent probes for in situ hybridization. J. Am. Chem.
Soc. 123, 4103–4104 (2001).
48. Medintz, I. L. et al. Self-assembled nanoscale biosensors based on quantum
dot FRET donors. Nature Mater. 2, 630–638 (2003).
49. Akerman, M. E., Chan, W. C. W., Laakkonen, P., Bhatia, S. N. & Ruoslahti, E.
Nanocrystal targeting in vivo. Proc. Natl Acad. Sci. 99, 12617–12621 (2002).
50. Slocik, J. M., Moore, J. T. & Wright, D. W. Monoclonal antibody recognition of
histidine-rich peptide encapsulated nanoclusters. Nano Lett. 2, 169–173 (2002).
51. Hainfeld, J. F., Liu, W., Halsey, C., M. R., Freimuth, P. & Powell, R., D. Ni-
NTAgold clusters target His-tagged proteins. J. Stuct. Biol. 127, 185–198
(1999).
52. Clapp, A. R. et al. Fluorescence resonance energy transfer between quantum
dot donors and dye-labeled protein acceptors. J. Am. Chem. Soc. 126, 301–310
(2004).
53. Medintz, I., L. et al. A fl uorescence resonance energy transfer derived structure
of a quantum dot-protein bioconjugate nanoassembly. Proc. Natl Acad. Sci.
101, 9612–9617 (2004).
54. Medintz, I. L., Trammell, S. A., Mattoussi, H. & Mauro, J. M. Reversible
modulation of quantum dot photoluminescence using a protein-bound
photochromic fl uorescence resonance energy transfer acceptor. J. Am. Chem.
Soc. 126, 30–31 (2004).
nmat1390-print.indd 445 11/5/05 10:18:22 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group
REVIEW ARTICLE
446 nature materials | VOL 4 | JUNE 2005 | www.nature.com/naturematerials
55. Goldman, E. R. et al. Conjugation of luminescent quantum dots with
antibodies using an engineered adaptor protein to provide new reagents for
fl uoroimmunoassays. Anal. Chem. 74, 841–847 (2002).
56. Hanaki, K. et al. Semiconductor quantum dot/albumin complex is a long-life
and highly photostable endosome marker. Biochem. Biophys. Res. Comm. 302,
496–501 (2003).
57. Sukhanova, A. et al. Biocompatible fl uorescent nanocrystals for
immunolabeling of membrane proteins and cells. Anal. Biochem. 324, 60–67
(2004).
58. Jaiswal, J. K., Mattoussi, H., Mauro, J. M. & Simon, S. M. Long-term multiple
color imaging of live cells using quantum dot bioconjugates. Nature
Biotechnol. 21, 47–51 (2003).
59. Kaul, Z. et al. Mortalin imaging in normal and cancer cells with quantum dot
immuno-conjugates. Cell Res. 13, 503–507 (2003).
60. Hoshino, A., Hanaki, K.-I., Suzuki, K. & Yamamoto, K. Applications of
Tlymphoma labeled with fl uorescent quantum dots to cell tracing markers in
mouse body. Biochem. Biophys. Res. Comm. 314, 46–53 (2004).
61. Chen, F. & Gerion, D. Fluorescent CdSe/ZnS nanocrystal-peptide conjugates
for long-term, nontoxic imaging and nuclear targeting in living cells. Nano
Lett. 4, 1827–1832 (2004).
62. Derfus, A. M., Chan, W. C. W. & Bhatia, S. N. Intracellular delivery of quantum
dots for live cell labeling and organelle tracking. Adv. Mater. 16, 961–966
(2004).
63. Voura, E. B., Jaiswal, J. K., Mattoussi, H. & Simon, S. M. Tracking early
metastatic progression with quantum dots and emission scanning microscopy.
Nature Med. 10, 993–998 (2004).
64. Derfus, A. M., Chan, W. C. W. & Bhatia, S. N. Probing the cytotoxicity of
semiconductor quantum dots. Nano Lett. 4, 11–18 (2004).
65. Pellegrino, T. et al. Quantum dot-based cell motility assay. Differentiation 71,
542–548 (2003).
66. Dahan, M. et al. Diffusion dynamics of glycine receptors revealed by
singlequantum dot tracking. Science 302, 442–445 (2003).
67. Lidke, D. S. et al. Quantum dot ligands provide new insights into erbB/HER
receptor-mediated signal transduction. Nature Biotechnol. 22, 198–203 (2004).
68. Levene, M. J., Dombeck, D. A., Kasischke, K. A., Molloy, R. P. & Webb, W. W.
In vivo multiphoton microscopy of deep brain tissue. J. Neurophys. 91,
1908–1912 (2004).
69. Rosenthal, S. J. et al. Targeting cell surface receptors with ligand-conjugated
nanocrystals. J. Am. Chem. Soc. 124, 4586–4594 (2002).
70. Kim, S. et al. Near-infrared fl uorescent type II quantum dots for sentinel
lymph node mapping. Nature Biotechnol. 22, 93–97 (2004).
71. Larson, D. R. et al. Water-soluble quantum dots for multiphoton fl uorescence
imaging in vivo. Science 300, 1434–1437 (2003).
72. Colvin, V. L. The potential environmental impact of engineered nanomaterials.
Nature Biotechnol. 21, 1166–1170 (2003).
73. Hoet, P. H., Bruske-Hohlfeld, I. & Salata, O. V. Nanoparticles - known and
unknown health risks. J. Nanobiotechnol. 2, 2–12 (2004).
74. Green, M. & Howman, E. Semiconductor quantum dots and free radical
induced DNA nicking. Chem. Comm. 121–123 (2005).
75. Kirchner, C. et al. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles.
Nano Lett. 5, 331–338 (2005).
76. Xiao, Y. & Barker, P. E. Semiconductor nanocrystal probes for human
metaphase chromosomes. Nucl. Acids Res. 32, 3 e28 (2004).
77. Gerion, D. et al. Room-temperature single-nucleotide polymorphism and
multiallele DNA detection using fl uorescent nanocrystals and microarrays.
Anal. Chem. 75, 4766–4772 (2003).
78. Gerion, D. et al. Sorting fl uorescent nanocrystals with DNA. J. Am. Chem. Soc.
124, 7070–7074 (2002).
79. Goldman, E. R. et al. Multiplexed toxin analysis using four colors of quantum
dot fl uororeagents. Anal. Chem. 76, 684–688 (2004).
80. Mamedova, N. N., Kotov, N. A., Rogach, A. L. & Studer, J. Albumin-CdTe
nanoparticle bioconjugates: preparation, structure, and interunit energy
transfer with antenna effect. Nano Lett. 1, 281–286 (2001).
81. Tran, P. T., Goldman, E. R., Anderson, G. P., Mauro, J. M., & Mattoussi, H. Use
of luminescent CdSe-ZnS nanocrystal bioconjugates in quantum dot-based
nanosensors. Phys. Status Solidi B 229, 427–432 (2002).
82. Clapp, A. R., Medintz, I. L., Fisher, B. R., Anderson, G. P. & Mattoussi, H. Can
luminescent quantum dots be effi cient energy acceptors with organic dye
donors. J. Am. Chem. Soc. 127, 1242–1250 (2005).
83. Samia, A. C. S., Chen, X. & Burda, C. Semiconductor quantum dots for
photodynamic therapy. J. Am. Chem. Soc. 125, 15736–15737 (2003).
84. Bakalova, R., Ohba, H., Zhelev, Z., Ishikawa, M. & Baba, Y. Quantum dots as
photosensitizers. Nature Biotechnol. 22, 1360–1361 (2004).
85. Xu, H. X. et al. Multiplexed SNP genotyping using the Qbead (TM) system:
a quantum dot-encoded microsphere-based assay. Nucl. Acids Res. 31, 8 e43
(2003).
86. Josephson, L., Kircher, M. F., Mahmood, U., Tang, Y. & Weissleder, R.
Nearinfrared fl uorescent nanoparticles as combined MR/optical imaging
probes. Bioconj. Chem. 13, 554–560 (2002).
87. Hohng, S. & Ha, T. Near-complete suppression of quantum dot blinking in
ambient conditions. J. Am. Chem. Soc. 126, 1324–1325 (2004).
88. Hu, J. et al. Linearly polarized emission from colloidal semiconductor
quantum rods. Science 292, 2060–2063 (2001).
89. Han, M., Gao, X., Su, J. Z. & Nie, S. Quantum-dot-tagged microbeads for
multiplexed optical coding of biomolecules. Nature Biotechnol. 19, 631–635
(2001).
90. Katz, E. & Willner, I. Integrated nanoparticle-biomolecule hybrid systems:
synthesis, properties, applications. Angew. Chem. Int. Edn Eng. 43, 6042–6108
(2004).
91. Ishii, D. et al. Chaperonin-mediated stabilization and ATP-triggered release of
semiconductor nanoparticles. Nature 423, 628–632 (2003).
92. Wang, D., He, J., Rosenzweig, N. & Rosenzweig, Z. Superparamagnetic Fe2O3
beads-CdSe/ZnS quantum dots core-shell nanocomposite particles for cell
separation. Nano Lett. 4, 409–413 (2004).
93. Mansson, A. et al. In vitro sliding of actin fi laments labelled with single
quantum dots. Biochem. Biophys. Res. Comm. 314, 529–534 (2004).
94. Bachand, G. D. et al. Assembly and transport of nanocrystal CdSe quantum
dot nanocomposites using microtubules and kinesin motor proteins. Nano
Lett. 4, 817–821 (2004).
95. Kloepfer, J. A. et al. Quantum dots as strain- and metabolism-specifi c
microbiological labels. App. Environ. Microbiol. 69, 4205–4213 (2003).
96. Patolsky, F. et al. Lighting-up the dynamics of telomerization and DNA replication
by CdSe-ZnS quantum dots. J. Am. Chem. Soc. 125, 13918–13919 (2003).
97. Menzel, E. R. et al. Photoluminescent semiconductor nanocrystals for
fi ngerprint detection. J. Forens. Sci. 45, 545–551 (2000).
98. Baird, G. S., Zacharias, D. A. & Tsien, R. Y. Biochemistry, mutagenesis, and
oligomerization of DsRed, a red fl uorescent protein from coral. Proc. Natl
Acad. Sci. 97, 11984–11989 (2000).
99. Michalet, X. et al. Quantum dots for live cells, in vivo imaging, and
diagnostics. Science 307, 538–544 (2005).
Acknowledgements
The authors acknowledge the Naval Research Laboratory (NRL) and A. Ervin and
L. Chrisey at the Offi ce of Naval Research (ONR grant N001404WX20270) and A.
Krishan at DARPA for support. I.L.M. was and H.T.U. is supported by a National
Research Council Fellowship through NRL.
Correspondence should be addressed to I.M. or H.M.
Competing interests statement
The authors declare that they have no competing fi nancial interests.
nmat1390-print.indd 446 11/5/05 10:18:23 am
Nature Publishing Group© 2005
© 2005 Nature Publishing Group缩略图:
当前页面二维码




 工程招标采购
工程招标采购 搞笑表情
搞笑表情 微信头像
微信头像 美女图片
美女图片 APP小游戏
APP小游戏 PPT模板
PPT模板